BiV Geometry¶
In this demo we will show how you can generate fibers using the ldrb algorithm on a BiV mesh. In order to run this demo you also need to install mshr if you haven’t already.
To run the demo in serial do
python demo_biv.py
If you want to run the demo in parallel you should first comment out the lines that don’t work in serial. Say you want to run on 4 cpu’s, you run the command:
mpirun -n 4 python demo_biv.py
import dolfin as df
import ldrb
# Here we just create a lv mesh. Here you can use yor own mesh instead.
geometry = ldrb.create_biv_mesh()
#
# The mesh
mesh = geometry.mesh
# The facet function (function with marking for the boundaries)
ffun = geometry.ffun
# A dictionary with keys and values for the markers
markers = geometry.markers
# Also if you want to to this demo in parallell you should create the mesh
# in serial and save it to e.g xml
# df.File('biv_mesh.xml') << mesh
# And when you run the code in paralall you should load the mesh from the file.
# mesh = df.Mesh('biv_mesh.xml')
# Since the markers are the default markers and the facet function is
# stored within the mesh itself, you can just set
# markers = None
# ffun = None
# Decide on the angles you want to use
angles = dict(alpha_endo_lv=30, # Fiber angle on the LV endocardium
alpha_epi_lv=-30, # Fiber angle on the LV epicardium
beta_endo_lv=0, # Sheet angle on the LV endocardium
beta_epi_lv=0, # Sheet angle on the LV epicardium
alpha_endo_sept=60, # Fiber angle on the Septum endocardium
alpha_epi_sept=-60, # Fiber angle on the Septum epicardium
beta_endo_sept=0, # Sheet angle on the Septum endocardium
beta_epi_sept=0, # Sheet angle on the Septum epicardium
alpha_endo_rv=80, # Fiber angle on the RV endocardium
alpha_epi_rv=-80, # Fiber angle on the RV epicardium
beta_endo_rv=0, # Sheet angle on the RV endocardium
beta_epi_rv=0) # Sheet angle on the RV epicardium
# Choose space for the fiber fields.
# This is a string on the form {family}_{degree}
fiber_space = 'Quadrature_2'
# fiber_space = 'Lagrange_1'
# Compute the microstructure
fiber, sheet, sheet_normal = ldrb.dolfin_ldrb(mesh=mesh,
fiber_space=fiber_space,
ffun=ffun,
markers=markers,
log_level=df.debug,
**angles)
# # Store the results
df.File('biv_fiber.xml') << fiber
df.File('biv_sheet.xml') << sheet
df.File('biv_sheet_normal.xml') << sheet_normal
# If you run in parallel you should skip the visualization step and do that in
# serial in stead. In that case you can read the the functions from the xml
# Using the following code
# V = ldrb.space_from_string(fiber_space, mesh, dim=3)
# fiber = df.Function(V, 'biv_fiber.xml')
# sheet = df.Function(V, 'biv_sheet.xml')
# sheet_normal = df.Function(V, 'biv_sheet_normal.xml')
# Store files in XDMF to be visualized in Paraview
# (These function are not tested in parallel)
ldrb.fiber_to_xdmf(fiber, 'biv_fiber')
ldrb.fiber_to_xdmf(sheet, 'biv_sheet')
ldrb.fiber_to_xdmf(sheet_normal, 'biv_sheet_normal')
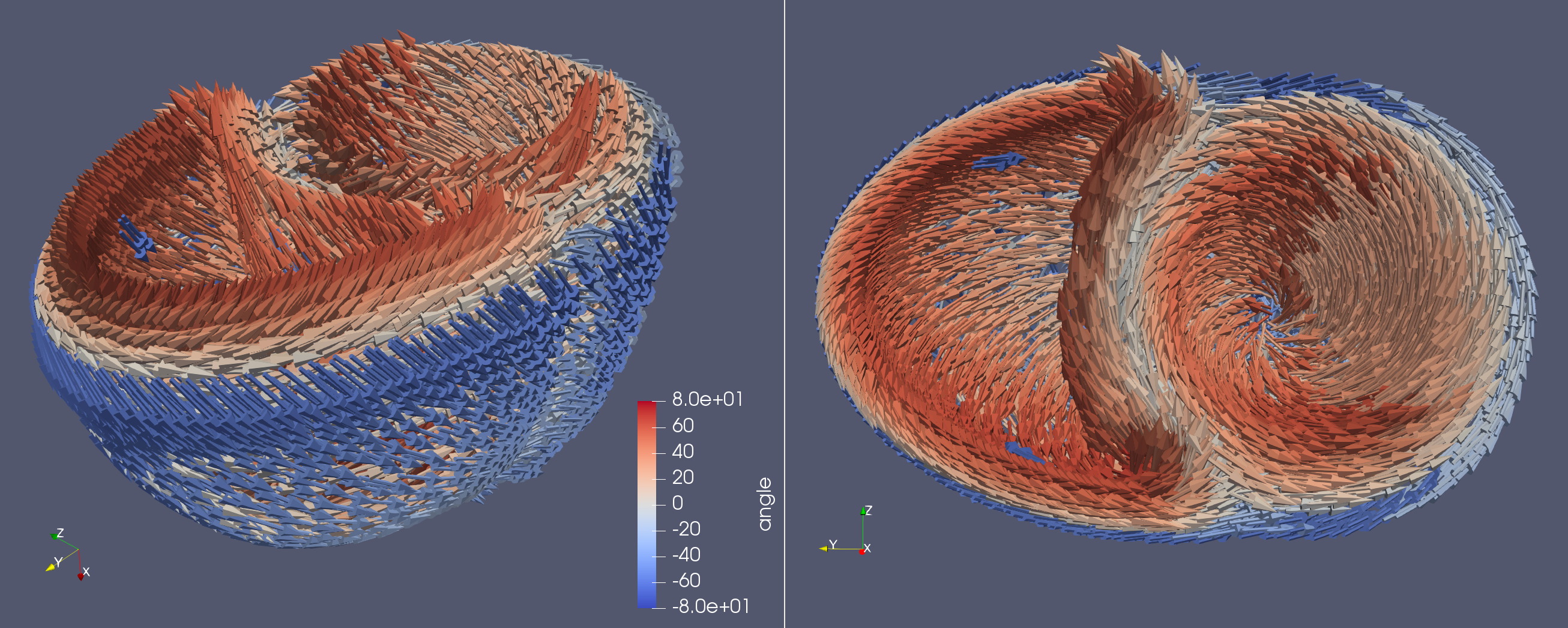
BiV Fiber